MemVerge has announced its BigMemory Optane-boosted DRAM system delivered a 25X increase in genomic sequencing speed at Analytical BioSciences.
Analytical BioSciences (ABio) is a single cell genomics company focusing on accelerating the development of therapeutics with faster sequencing. Genomic sequencing is used to identify virus strains such as SARS-CoV-2 variants, and computing a single-cell RNA sequence can take many hours.
The process is compute-intensive and entails many stages. For instance, large matrices need to fit in the server memory; intermediate stage results are saved and then reloaded for other stages. This introduces storage and recovery bottlenecks that are exacerbated by stage repetition for parameter tuning. However, overall execution time drops by more than half when Optane Persistent Memory is used to increase overall memory capacity.
Chris Kang, Head of Bioinformatics Operations at ABio, issued a statement: “The Big Memory platform that MemVerge and Intel developed accelerates our workflows and helps us generate results much faster, which will lead to more efficient ways to gain greater insights and knowledge in diseases mechanisms and improve healthcare.”
Benchmarks
MemVerge and ABio compared single-cell sequencing runs on a server incorporating two Xeon Gold 18-core CPUS and 192GB of DRAM, and the same server with equipped with with MemVerge’s MemoryMachine software plus 1,536GB of Optane DIMMs.
MemoryMachine combines DRAM and Optane into a single memory pool. The matrix used for the test run was 31,787 x 813,348 cells in size and we have charted and tabulated the results using data from the MemVerge case study
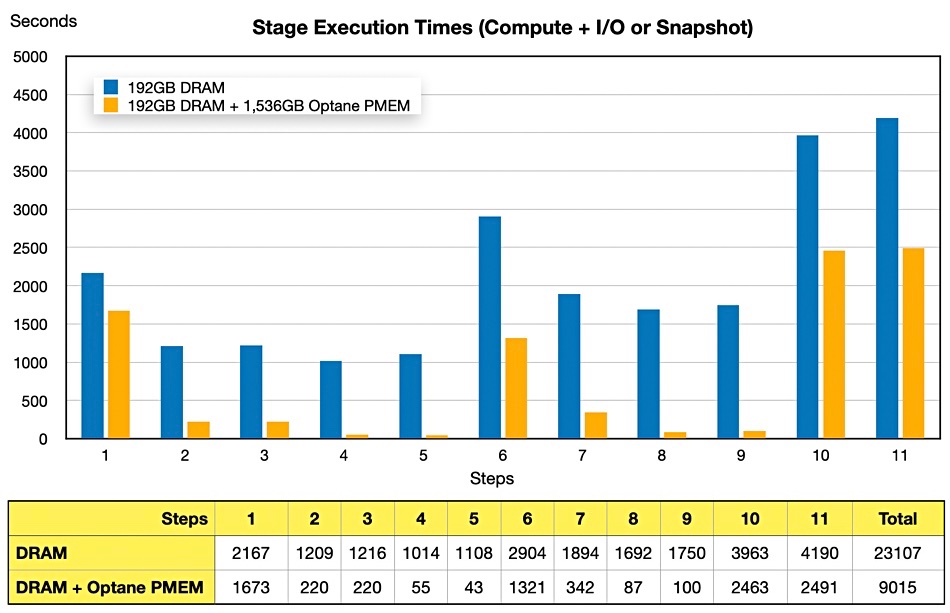
The intermediate stage result data is stored and accessed in the Optane memory pool, instead of slow access storage drives, which means that overall and stage execution times are reduced. Step 5 (above) shows the greatest improvement, with the Optane-boosted system completing more than 25 times faster.
The overall execution time using only DRAM was 23,107 seconds (6.4 hours) and the Optane using server completed the task in 9,015 seconds (2.5 hours), a 60 per cent reduction. The case study does not say what storage was used by the server i.e. whether it was SSD (fast) or hard disk drives (slow).
Slow disk drive storage increases the DRAM-only execution times due the relatively long time it takes to write intermediate results to the disks and then read them back.
MemVerge CEO Charles Fan said: “Until now, memory infrastructure did not offer a viable alternative to storage for genomic sequencing. Big Memory offers the same high-performance as DRAM at a dramatically lower cost, and with the persistence and agility needed for complex data pipelines.”
By extension, other bioinformatics analyses that use large matrices derived from next-generation sequencing techniques can also be accelerated.








