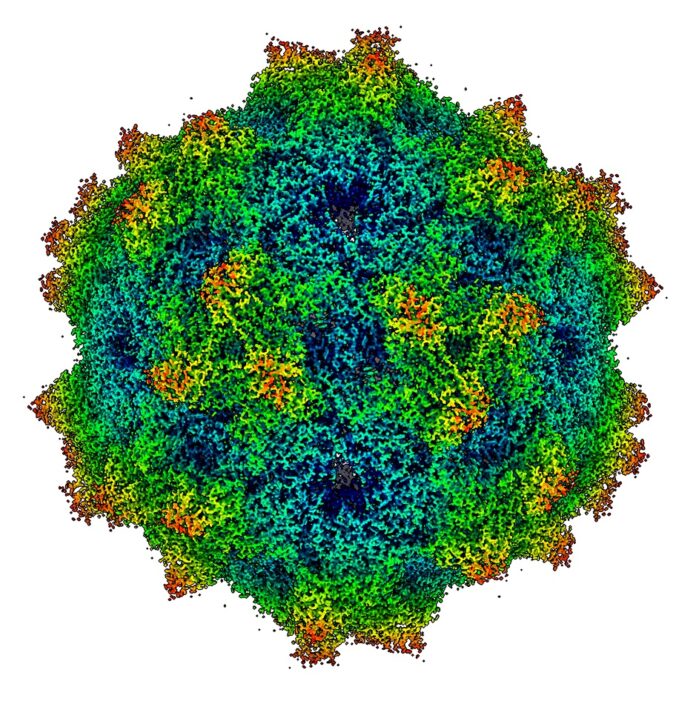
Case study. Oregon Health and Science University (OHSU) researchers in Portland are using cryogenically cooled electron microscopes to build 3D images of biological molecules as they investigate how proteins affect the brain, the COVID virus, serotonin neural processes, ageing and the myriad other aspects of human and other organisms’ biology.
Data from the instruments are stored in a Quobyte file system – chosen when OHSU decided it needed its own local storage rather than relying on an external system provided by the Pacific Northwest National Laboratory (PNNL). The storage system has to support high-performance computing applications used by researchers as well as being a vault for the instrument-generated data.
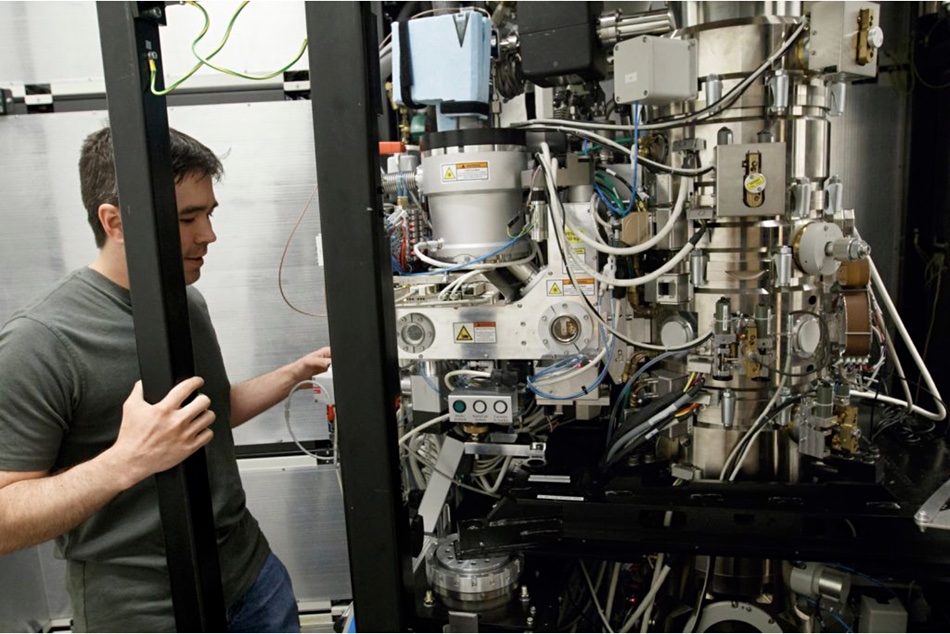
The OHSU Cryo-EM center has four telephone kiosk-size Titan Krios 300-kiloelectron volt Transmission Electron Microscopes (TEM) to visualize proteins and other biological molecules in 3D and at near-atomic scale. They examine samples cooled down to liquid nitrogen temperature and use high-energy electrons to view their structures down to a resolution of 2.8 Å.
Each Krios has a direct-electron detection camera fitted, and is mounted on a vibration absorbing pad inside its booth to keep its detectors as still as possible. Without this, they would be affected by vibrations as small as that caused by a person’s voice, a breeze, or currents in the adjacent Willamette river.
OHSU also has other microscopes, such as a Glacios Cryo-TEM, to pre-screen samples before using a Krios to get near-atomic resolution of selected images. Using a multi-million dollar Krios for pre-screening is overkill.
Each Krios generates an average 3TB of data a day, meaning 8 to 16TB/day for OHSU’s cryo-EM facility. We’re looking at around 120TB/week, up to 6.2PB/year, and that data has to be kept for use in the following 12 months or so. A proportion of it may be noise rather than signal, but the researchers may be able to pull out more signals from the noise with better algorithms in the future. So the data is kept.
There are 900 researchers using the cryo-EMs and accessing the data with up to 200 active projects at any given time.
Craig Yoshioka, a PhD and Research Associate Professor at OHSU directs its cryo-EM center. He said the original storage system was based on ZFS running on Linux servers. This could accept the data from the instruments fine, but could be slow at delivering it to the HPC apps.
The possible options he considered for fixing the slowness included simply scaling it up, or moving to a BeeGFS alternative, a Panasas system, a Quobyte cluster setup, VAST Data array, or a WekaIO file system. He specifically wanted a distributed file system accessed via a centralized web interface with a single namespace and intuitive management utilities.
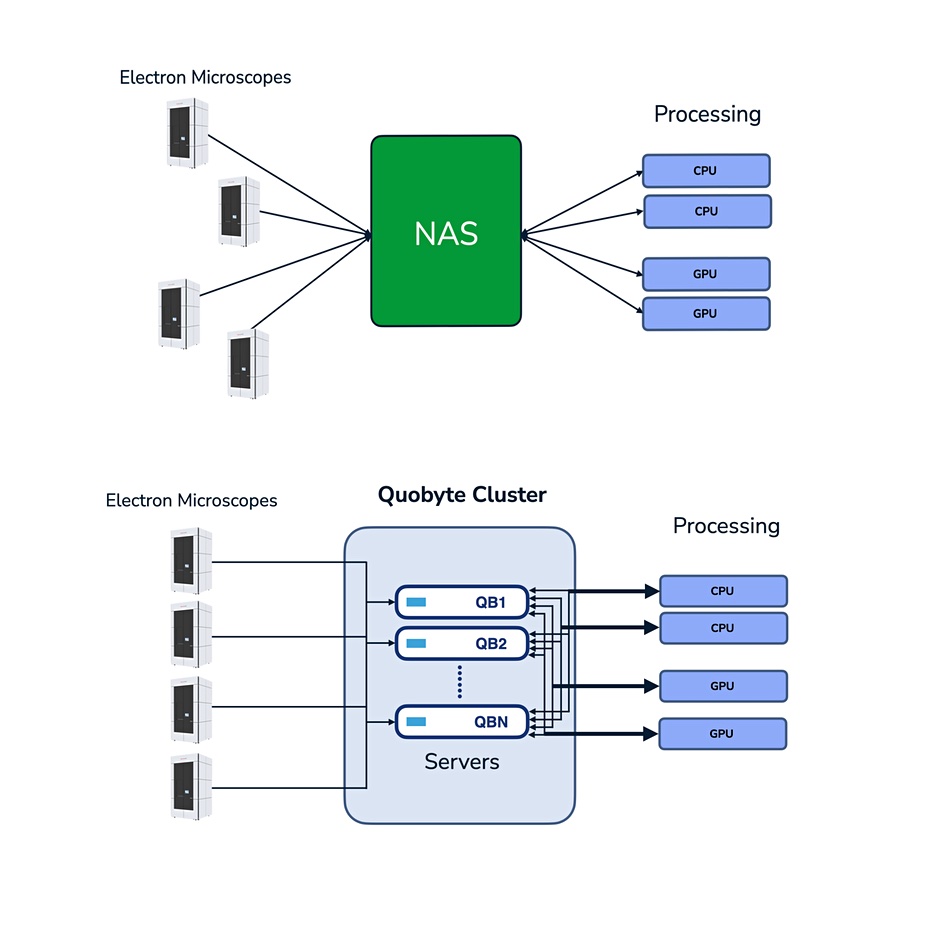
He inspected Quobyte in November last year, liked what he saw, and decided to use its software running on a mix of hard disk and solid state drives. His team had migrated and loaded 1.5PB of data onto the Quobyte system by January this year and the system is working just fine.
Bootnote
Quobyte is doing well in Oregon. Another customer there is The Center for Quantitative Life Sciences (CQLS) at Oregon State University.