Chinese scientists in Tianjin University have stored and retrieved 445KB of digital data from living E. coli bacteria cells and say E.coli represents a stable DNA storage medium.
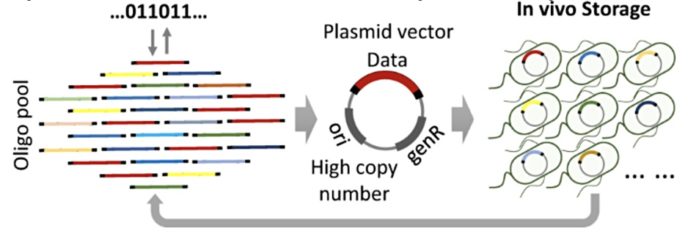
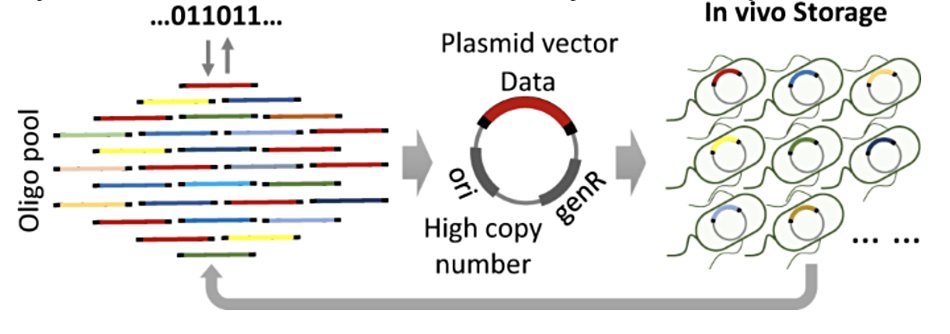
Up until now, digital data storage in DNA has used synthetic DNA stored in glass phials or similar containers. A Chinese team of research scientists has done it by inserting synthetic DNA into living cells and then retrieving the data after the cells have reproduced.
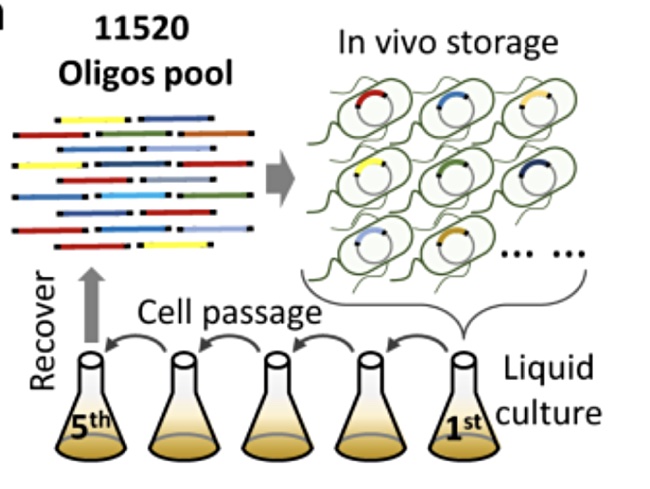
The researchers said artificially stored DNA, stored in vitro or glass, typically use short lengths of DNA with fragment lengths ranging from 100 to around 200 nucleotides. Their method, stored in vivo (living cells) uses much larger fragments, with up to 11,520 nucleotides.
As the cells reproduce they make new cells which carry the inserted digital data-carrying DNA fragments. The researchers said “the genomic maintenance mechanism of living cells ensures that the DNA molecules are replicated with high fidelity.” This means the fragments can be retrieved from a larger number of cells than the starting population, meaning “higher stability and longer storage periods could be expected.”
The details are in a Nature paper and the researchers looked at oligos, short for oligonucleotides, which are single short strands of DNA. They used more than 10,000 of them encompassing 2,304 kilo bit pairs (kbps). This is the unit of length measurement for DNA equal to 1,000 base pairs. A base pair is two of the four DNA nucleobases combined, meaning adenine with thymine, and cytosine with guanine.
The source binary data was encoded into the 4-letter DNA alphabet formed from the four nucleobases; A, C, G and T. There was an encoding redundancy of 1.56 per cent at the software level to tolerate the physical loss of some oligos. They built oligo pools of up to 11,520 distinct oligos. These were assembled into plasmid vectors, twin-stranded circular DNA molecules which replicate independently within a cell. These were assembled in a redundant fashion and stored in a mixed E. Coli culture on solid plates or in liquid medium. There, the cells reproduced and formed colonies.
This number of oligos remained stable in the mixed culture of E. coli cells even over multiple divide and split operations in which a culture in one medium was divided into two, left to grow again, and then divided once more. Up to five such splits, “passages” in the scientists’ terminology, were demonstrated.
The cell’s digital data contents were read via DNA sequencing and found to be correct. The reading process involved plasmids carrying the digital information being isolated from a large liquid culture. Then a large number of oligos was recovered following a digestion process. There was little contamination from the host cell’s own DNA and that was removed using bio-reagents.
In summary the researchers concluded: “DNA storage inside cells has distinct advantage in terms of stable DNA maintenance for long periods of time and very low cost of replication.”
They claimed their work “is the largest scale archival data storage in living cells reported so far, paving the way for biological data storage taking advantage of both in vitro synthesis capacity and the biological power of living cells in an economical and efficient way, which is crucial for developing practical cold data storage on a large scale.”
The paper is entitled “A mixed culture of bacterial cells enables an economic DNA storage on a large scale” published in Communications Biology, volume 3, Article number: 416 (2020). It is credited to Min Hao, Hongyan Qiao, Yanmin Gao, Zhaoguan Wang, Xin Qiao, Xin Chen & Hao Qi.