A DNA-based data storage system can store millions of times more data in the same volume as a conventional system, lasts for thousands of years at room temperatures, and gives you the ability to physically own and easily transport your data.
Current DNA storage technologies are costly and slow. For example, University of Washington and Microsoft researchers described an automated DNA storage proof of concept in a March 2019 Nature Scientific Reports paper. They encoded data, the word “hello” in DNA, stored it and then read it back. Their paper says the “system’s write-to-read latency is approximately 21 hours.
The race is now on to build a commercially viable DNA data storage technology, with Microsoft, Georgia Tech and several startups including Catalog Technologies, Iridia, and Helixworks Technologies throwing their hats into the ring.
In this article we take a closer look at Catalog Technologies, an MIT spinoff, which declares DNA storage has been prohibitively slow and expensive, until now. It claims it is making DNA data storage economically feasible for the first time. Catalog is tiny but thinks it has stolen a march on its competitors. “We’re positioned to make this a reality within the next year or two, rather than in five or six years,” CEO Hyunjun Park told MIT in 2019.
The Boston-based startup was founded in 2016 by Park, a microbiologist, and Nathaniel Roquet, chief technology and innovation officer, who is a biophysicist. The company emerged from stealth in June 2018 and has received $10.5m in funding, according to the 2019 MIT article.
The ABC of DNA
Catalog’s ultimate goal is to offer DNA data storage-as-a-service to customers that need to store petabytes of data in archives.
It is still early days but the company thinks DNA storage will be less expensive than on-premises tape libraries and cloud storage services. It thinks it can get DNA storage costs down to under a three thousandth of a cent per MB.
Catalog is developing a DNA-based data storage system to hold and process massive amounts of data. As a demo of DNA as a medium for ultra-long term archival of data, Catalog has encoded the entire contents of Wikipedia – about 16GB of compressed data – into DNA.
In June 2019 Park told his alma mater MIT that Catalog was readying a demonstration system.

In a 2019 video Park explains that Catalog treats DNA as an alphabet.
“Catalog’s method is to synthesize batches of a bunch of different kinds of short DNA sequences, which can be thought of as analogous to letters. The original binary data is encoded by stitching together these DNA letters into billions of possible words.”
The short sequences are about 30 to 40 base pairs long; combinations of the As, Gs, Cs, and Ts of the genetic code, and there are around 200 of them.
The resulting less than a millilitre of fluid, or dry powder pellets, containing the DNA molecules, can remain stable for thousands of years, according to Park. It can be sequenced back [read] whenever information retrieval is needed.
Writing speed
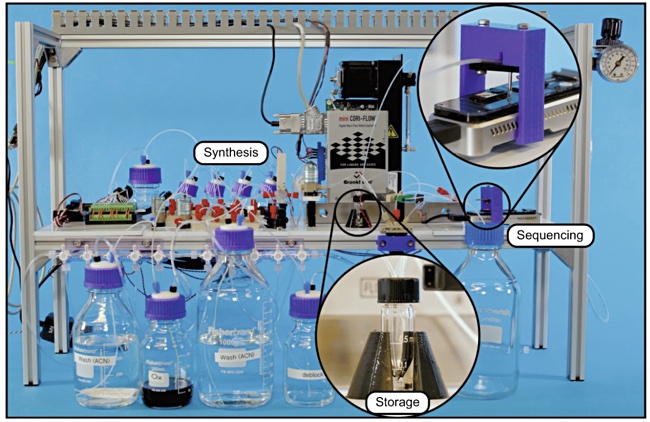
Working with UK-based Cambridge Consultants, Catalog proved the feasibility of using its proprietary DNA data encoding method in a test machine or instrument. This recorded 1Mbit of data per 24-hour day into DNA.
Catalog compares its machine to a printing press with movable typefaces. These typefaces are pre-built DNA molecules in different combinations, and this obviates the need to synthesize billions of different molecules.
A Mbit/day rate is better than the 5bytes in 21 hours demonstrated by the University of Washington and Microsoft researchers, but Catalog reckoned it needed to be a million times faster than that; 1Tbit per day instead of 1Mbit per day.
In October 2018 it announced it was working again with Cambridge Consultants on building a terabit writing instrument that would encode DNA “for about 1/1,000,000 the cost of what’s been possible before.”
Park said at the time: “The machine we are developing with Cambridge Consultants will bring DNA data storage out of the research lab and into the real world, for the first time in history.”
He thinks this leap in speed will help make it economically attractive to use DNA as the medium for long-term archival of data.
That is a great advance in DNA storage writing. But it is slow in data storage write bandwidth terms. For instance, a 14TB WD Purple disk drive spinning at 7,200rpm reads and writes data at up to 255MB/sec.This is 176 times faster than Catalog’s prototype.
The upfront trade-off is that you write data 176 times slower than a disk drive to store it thousands of times longer in an offline way. This is analogous to a magnetic tape cartridge but in a much smaller space.

DNA dot printer
The terabit writer prints – quasi-inkjet style – DNA ‘letters’ into dots on a sheet of plastic film. A dot corresponds to a block of binary data and contains multiple DNA letters. These are fixed through drying and then washed off the film into a fluid which is turned into pellets for storing.
The pellets are rehydrated and fed through a genome sequencer when the information they hold has to be retrieved.

Reading DNA storage
Writing data into DNA storage is one thing. Reading it is another. Assume you have some store of pellets holding DNA molecules representing encoded digital information. How do you read them?
We have asked Park directly about the reading process but he has not replied. Cambridge Consultants declined to comment.
In the absence of definitive information Blocks & Files speculates this will involve a robotic library with automated movement of desired pellets to a ‘drive’, which rehydrates them and feeds them into a DNA sequencer. It then reads a pellet’s contents and outputs a file of digital data. A tape library is the analogy we have in mind.
We are interested in finding out if this is, as it seems, a destructive read, or if the pellet can be recreated and moved back to its location in the library.
DNA computing’s far out future
According to Catalog, DNA storage is nothing new … we humans have been using it for millions of years.
But in common with the University of Washington and Microsoft researchers, Catalog conceives a future for DNA computing. This is the notion of using data stored in DNA molecules and manipulating it directly, using enzymes for example. This would be performed in a highly parallel way, with potentially millions of simultaneous operations.
It all seems very far away at this stage. Watch the video above for more information on this notion of a way to bring computation to storage.
We will explore Iridia, Helixworks and Microsoft’s efforts with DNA data storage in subsequent articles.