Research scientists have demonstrated kilobyte-scale data storage in synthetic metabolitic molecules, smaller than DNA, with data read by mass spectrometry. This could lead to dense offline storage that may be denser than tape reels or flash memory.
Metabolites are intermediate chemical compounds created during metabolism, the chemical reactions inside organisms that sustain life, such as sugars, amino acids, nucleotides, vitamins and anti-oxidants. A metabolome is the set of such metabolites found within a biological sample.
Their presence or absence in an organic chemistry sample can be detected by mass spectrometry.
A recent demonstration of DNA-based data storage by researchers at Washington State and Microsoft inspired scientists at Brown University to explore the potential of the metabolome as a biomolecular information system.
Metabolite molecules that are smaller than DNA and thus denser than DNA storage.
Biomechanical storage medium
A team led by Jacob Rosenstein has published a paper demonstrating the writing and reading of kilobyte-scale images using synthetic metabolomes containing a set of 36 metabolites.
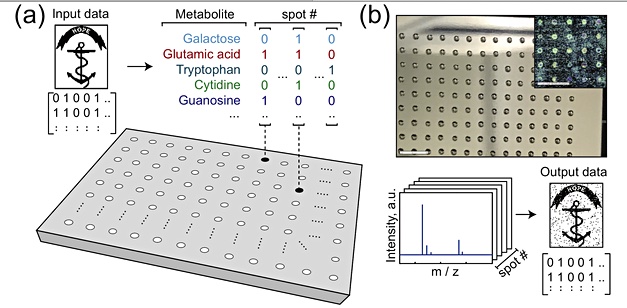
The presence or absence of a specific metabolite in a sample signals a binary one or zero and they can be grouped to provide, for example, 4-bit quartet, such as 1010. Each metabolite would be viewed as a bit position in this quarter and signify 1 or 0 in that position.
The samples were created as nanoliter-sized volumes or spots deposited in their thousands in a grid pattern on 76 x120 mm² steel plates, with spots deposited on a 48 x 32 grid; 1,536 spots in total.
An acoustic liquid handler was used for the deposition. Each spot contains a mixture of metabolites, obtained from a chemical library of purified metabolites. This library represents a synthetic metabolome.
The total number of bits stored in a single spot is given by the number of metabolites in the library. Four were used in the experiments described in the paper but 36 is the upper bound in this particular work.
The metabolites were carried in a solvent mixture, deposited on the plate and the solvent then evaporates. The plates were left to dry and the spot compounds to crystallise overnight.
The researchers used mass spectrometry to analyse the spots and determine the presence or absence of metabolites in each spot, thus building up the values of a 4-bit quartet at each spot. Each spot’s binary values were read in parallel, with the spots on the plate read serially. This took under two hours.
Statistical analyses were used to determine if a specific metabolite was present or absent at a spot and the accuracy reached was about 99.6 per cent.
Several experimental results were reported. One involved 1,024 spots containing 6 metabolites (sorbitol, glutamic acid, trypotophan, cytidine, guanosine and 2-deoxyguanosine hydrate) used to encode a 6,142 pixel image of an ibex. In effect, 6-bit words were used for the encoding.
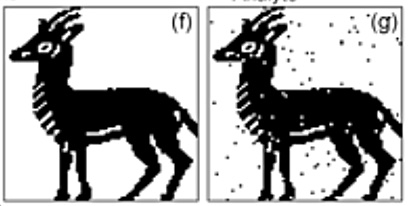
The ibex before (f) and after (g) images above show that the process is not that accurate, with random values dotting the image. The researchers observed a two per cent cumulative read/write error. Also repeated spot reading caused data loss, with a <1 per cent error added by each successive read.
A further experiment used a 17,424 pixel image of a cat, (i) below, which was encoded into 1,452 spots using 12 metabolites per spot, 12-bit words in effect.

Once again random errors were apparent in the read image (ii above) with better statistical analyses (multi-peak logistic regression) improving the read accuracy (iii).
Comment
The researchers successfully explored their stated aim to explore the proof of principle of the metabolome as a storage medium. But any practical application could be decades away.
The Brown researchers concluded that “the metabolome is a viable and robust medium for representing digital information.”
The medium is certainly viable but “robust” is a bit of stretch. Accuracy will need to improve.
The Brown researchers think they can improve on the density of DNA storage, currently 2.4PB/gram.
In their experimental demonstration they wrote data at 5bits/sec and had aggregate read speeds of 11 bits/sec.
Let’s be blunt: these read and write speeds are appallingly slow by disk drive and NAND flash standards.
According to the researchers, read speed improvements are possible through increased metabolite library size; meaning more bits per spot. They think hundreds of bits per spot could be achievable.
Write speed and density improvements would come from reducing the spot size. They wrote: “Scaling the mixture spots down to diffraction-limited laser spot scales could improve data storage density by six orders of magnitude. Theoretically, this could facilitate extension from kilobyte- to gigabyte-data sets per plate.”
Blocks & Files notes that M.2 2280 flash drives are 22mm x 80mm in size and already store up to 1TB (Optane H10 using QLC (4bits/cell) flash) with read and write rates of 2.4/1.8GB/sec.
Metabolomitic-based data storage would require a lot of development to match this.







