Chinese scientists have stored and retrieved data in DNA on a single electrode instead of using complicated chemical laboratory equipment for synthesising and manipulating DNA in the storage process. This is an enormous advance in DNA storage equipment size and convenience, yet the data rates are excruciatingly slow.
The Southeastern University research team’s paper was published in Science Advances and entitled “Electrochemical DNA synthesis and sequencing on a single electrode with scalability for integrated data storage.”
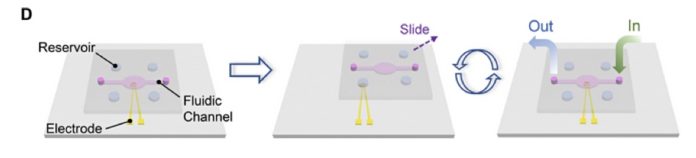
The paper’s abstract talks about “the synthesis and sequencing of DNA on a single electrode with scalability for an integrated DNA-based data storage system.” A key advance was using a so-called SlipChip, a microfluidic device to hold the DNA chemicals and the various reagents.
There are two plates or layers in a SlipChip, with the lower plate having holes or wells in it to hold the chemical liquids or microdroplets. The top plate acts as a lid for the lower plate’s wells but it also has its own wells and ducts. By moving or “slipping” the top plate one or more of its wells can be lined up via ducts with wells in in the bottom plate and liquids in the wells can then react via the ducts.
No pumps or valves are needed to move the chemicals and bring them into contact with each other. A single SlipChip can be an electrode and its electrical charge altered by the presence or absence of DNA sequences. The paper’s abstract states: “The synthesis of DNA is based on phosphoramidite chemistry and electrochemical deprotection. The sequencing relies on charge redistribution originated from polymerase-catalyzed primer extension, leading to a measurable current spike.”
The SlipChip device simplifies the liquid introduction involved in DNA synthesis and sequencing. This is because current DNA storage methods “usually involve complicated liquid manipulations in each step and manual operations in between. Adding one phosphoramidite nucleotide monomer in the synthesis step generally requires the introduction of at least four kinds of liquid solutions, not to mention the sequencing step. These limit the scale-up capability of this technique and increase the error probability.” Data error likelihood requires data redundancy — extra data in other words. Random access capabilities can mean adding address tags to the DNA sequences “which can become markedly cumbersome with scaling up” and contribute to data loss due to possible biased amplification.
The equipment needed is large, cumbersome and the process requires many steps, often manual and hence error-prone. The Chinese researchers effectively invented a DNA synthesis storage and retrieval system on a chip, a SlipChip, using a gold electrode.
Binary data was encoded into the quaternary (A, T, C and G) DNA base sequences which are then synthesised into DNA. A sequencing process is used to read the data. All of the liquid manipulations for the synthesis and sequencing are accomplished with the SlipChip.
The researchers wrote the Southeastern University motto — “Rest in the highest excellence!” — in the DNA and then read it back with acceptable accuracy: 87.22 per cent. Error correction based on data redundancy added in the encoding step served to achieve 100 per cent.
Here, for us at Blocks & Files, is the kicker: “It took about nine hours to write and read 4.5 bytes of data on the single electrode, which can be further improved by scaling up using an electrode array.” That’s 0.5 bytes/hour.
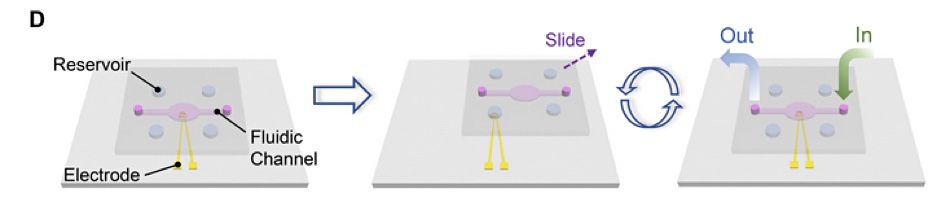
The researchers “developed a SlipChip-based microfluidic device for integrated DNA-based data storage. Briefly, a 2×2 Au electrode array was fabricated on a glass slide by standard photolithography and physical vapour deposition. A block of polydimethylsiloxane (PDMS) with a fluidic channel and reservoirs was fabricated and then assembled with the glass slide. The reservoirs were preloaded with reagents for DNA synthesis and sequencing on the electrode.”
The data writing process is fascinating. “To initiate the DNA synthesis, we slipped the top PDMS relative to the bottom glass slide to expose the electrodes to phosphoramidite nucleotide monomers in the reagent reservoirs. Then, washing, oxidation, and electrochemical deprotection steps were sequentially conducted by aligning the electrodes with corresponding reagent reservoirs and the fluidic channel, respectively.”
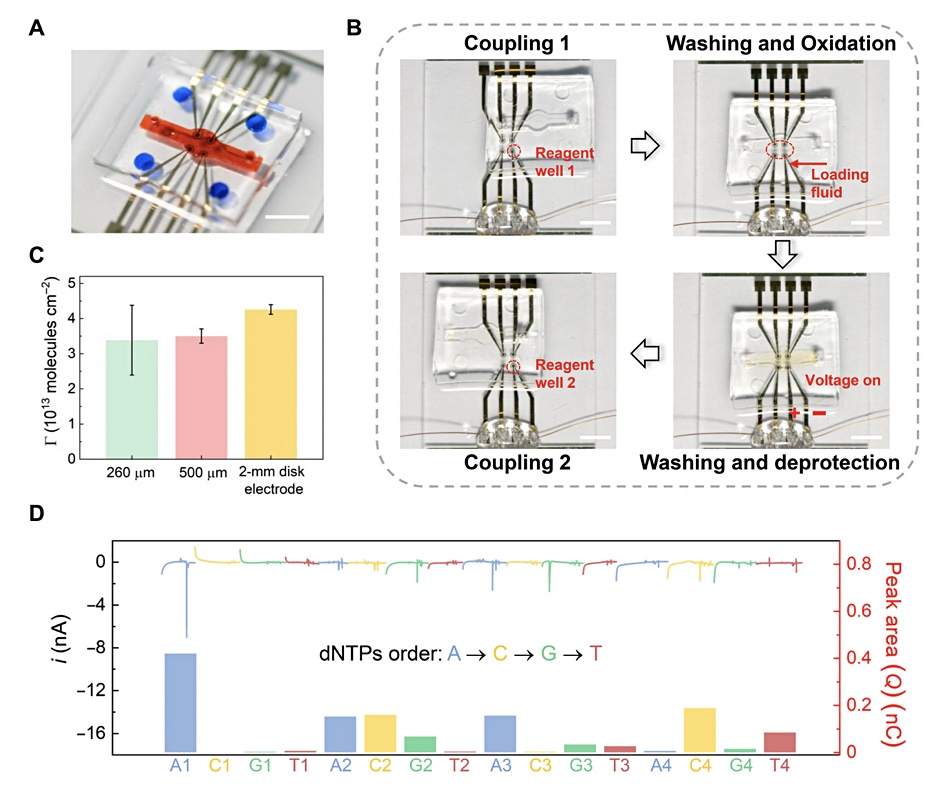
Reading the data via DNA synthesis involved deoxyribonucleoside triphosphate (dNTP). “Sequencing on the same electrode was accomplished by incubating the electrodes in the polymerase solution in the fluidic channel and then sequentially exposing the electrode to four reservoirs containing different dNTPs. During the process, the current from the electrode was measured for sequencing.”
Overall accuracy was 89.17 per cent, which was bumped up to 100 per cent by error correction through data redundancy again.
The researchers found it took about 14 hours to write and read 20 bytes of data on the four-electrode array. That’s 1.43 bytes/hour — better than 0.5 bytes/hour but still devastatingly slow.
Western Digital’s latest 20TB Gold disk drives transfer data at 269MB/sec — 269 million bytes a second. Uprate this to a per minute rate and then a per hour rate and we arrive at a speed that is 677 billion times faster than the SlipChip DNA storage process. If we start thinking about NAND storage, then a Toshiba FL6 writes data at 5,800 MB/sec. That means it’s upward of 14 trillion times faster than the SlipChip 4-electrode system.
It seems to us that, although DNA storage has potentially a fantastically high storage density — up to 455EB per gram — its speed is catastrophically slow compared to disk and worse still compared to NAND. So much so that it could well prevent DNA storage ever becoming a general commercial storage mechanism, despite its estimated million-year-plus lifespan.